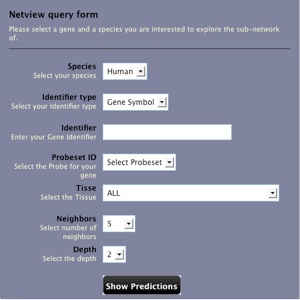
Netview how to...

Netview is a tool that allows to explore the co-expression network surrounding a gene of interest. The user can select one of the two species analyzed (i.e. human and mouse). The type of identifiers; probeset refers at the microarray model HG-U133A for human and Mouse430_2 for mouse species. If the user select a tissue, the visualization tool will provide a check-box to highlight those interactions that connect genes significantly expressed into the selected tissue.
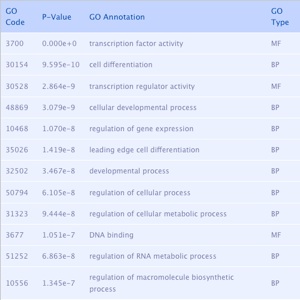
Output 1 (left table):
list of gene ontology terms that are significantly associates with the list of genes part of the explored sub-network.
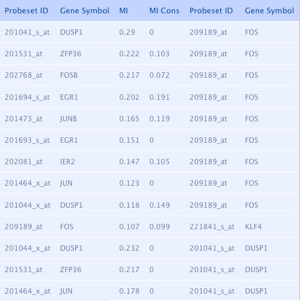
Output 2 (right table):
list of predicted interactions that are found to be statistically significant.

Netview button:
the user can graphically explore the sub-network via the jsquid java applet.